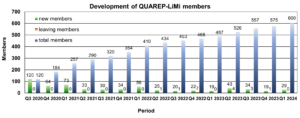
QUAREP-LiMi has reached another milestone. By the end of 2025 more than 700 members have joined QUAREP-LiMi. When we launched this initiative, we never imagined the community would grow to include so many members from 46 different countries around the globe. A heartfelt thank‑you to all our members for your continued support and contributions.
We are thrilled that WG 4 – System chromatic aberration and co-registration Power published its first protocol “Ensuring accurate co-registration measurement for quality control of Single Point Confocal Laser Scanning Microscopes – V1“. The protocol is featured on protocols.io
We warmly welcome our first member from Ukraine, Dr. Borys Olifirov, from the Bogomoletz Institute of Physiology of NAS of Ukraine who just joined Working Group 2, 5, 6, 7 and 8.
Figure 2: Entity-Relationship diagram comparing the the representation of an Objective hardware and acquisition settings in OME vs. NBO-Q.
In the absence of community Microscopy Metadata guidelines and tools, it is inherently difficult for manufacturers to incorporate standardized configuration information and performance metrics into image data and for scientists to produce comprehensive and harmonized records of imaging experiments. To address this challenge, in 2021, the NIH-funded 4D Nucleome (4DN) initiative Imaging Standards Working Group (IWG), working in conjunction with the BioImaging North America (BINA) Quality Control and Data Management Working Group (QC-DM-WG), proposed flexible microscopy metadata specifications for light microscopy
Join QUAREP WG7 (Metadata) LiMi-Model The Light-Microscopy (LiMi) Model aims to harmonize the description of light microscopy hardware, acquisition settings, and quality-control metrics to enhance image quality, reproducibility, and to fulfill the FAIR (Findable, Accessible, Interoperable, and Reusable) data principles. Microscopy metadata is essential for image data QC, interpretation, analysis and sharing. Purpose Promote the harmonized generation and pre-publication management of image datasets from the ground up. Facilitate the deposition of microscopy datasets to public image data repositories (e.g., BioImage
The LiMi-Model drives scientific progress Funders and journal editors can drive scientific rigor and transparency by endorsing the LiMi-Model, a community standard for capturing essential metadata in light microscopy. Supporting the LiMi-Model helps ensure imaging data is reproducible, FAIR-compliant, and suitable for long-term reuse and peer review.
The LiMi-Model empowers microscope users and imaging scientists! Microscope users and imaging scientists benefit from these standards that provide a clear, community-driven framework for recording the metadata that matters most—from sample preparation and imaging conditions to microscope hardware and acquisition settings. By adopting the LiMi-Model in your workflows, you ensure that your image data is not only reproducible and high quality but also compliant with FAIR principles, making it easier to share, publish, and reanalyze. The LiMi-Model community is actively
The LiMi-Model helps manufacturers and industry partners meet the community needs! Instrument manufacturers and industry partners can benefit from the LiMi-Model framework and definitions, allowing them to provide a consistent and comprehensive set of standardized metadata compliant with the community derived the LiMi-Model standards. This metadata complements any recorded image data helping data workflows and tools to process data and ensure data quality and consistency.
The LiMi-Model is built for software developers! The LiMi-Model provides a structured, community-agreed metadata framework for light microscopy, making it easier to build tools that ensure reproducibility, quality control, and interoperability. By aligning with the LiMi-Model, developers can create applications that integrate seamlessly with emerging standards like OME-Zarr and tools like Micro-Meta App, supporting FAIR data practices and accelerating scientific discovery.
Microscopy metadata is essential for image data QC, interpretation, analysis and sharing.
NBO-Q is for Developers Developers NBO-Q provides a structured, community-agreed metadata framework for light microscopy, making it easier to build tools that ensure reproducibility, quality control, and interoperability. By aligning with NBO-Q, developers can create applications that integrate seamlessly with emerging standards like OME-Zarr and tools like Micro-Meta App, supporting FAIR data practices and accelerating scientific discovery. Developers NBO-Q provides a structured, community-agreed metadata framework for light microscopy,making it easier to build tools that ensure reproducibility, quality control, and interoperability.
In April 2025, Yury Belyaev from the Microscopy Imaging Center (MIC) at the University of Bern, and a member of QUAREP-LiMi Working Group 5 (WG5), visited the Central Analytical Facilities (CAF) Microscopy Unit at Stellenbosch University, South Africa, as part of the Global BioImaging (GBI) International Job Shadowing Program During his visit, Yury Belyaev collaborated with Lize Engelbrecht, manager of the CAF Microscopy Unit, to organize a hands-on microscopy workshop. The training covered wide-field and confocal microscopy, image analysis, and